
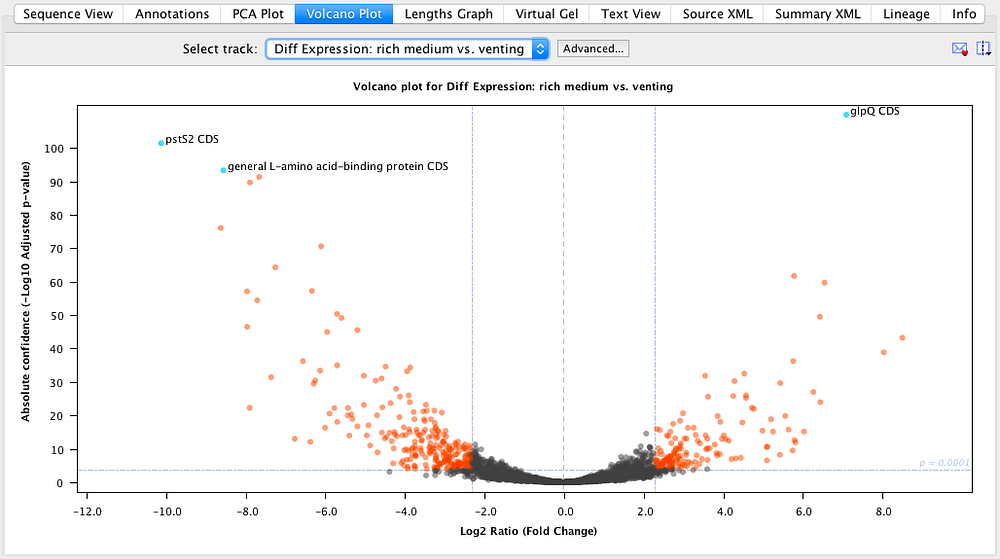
As of yet, no attempt has been reported to sequence and assemble the genome of A. acetabulum and its sister species dropped towards the end of the 1990s. crenulata, he observed that the cap developed into the morphology of the basal donor, demonstrating that the nucleus-containing rhizoid was in control of the morphogenetic fate of the cell 10, 11.ĭespite its popularity and importance in early cell biology and genetics, the interest in A. By transplanting and exchanging the apical and basal parts between A.
Already in the 1930s, Joachim Hämmerling used Acetabularia to prove that cellular morphogenesis was influenced by so-called “morphogenetic substances” (later confirmed to be RNA) which were produced by the nucleus and distributed to the rest of the cell 9. The size and highly elaborate cellular morphology, together with a large and distinct nucleus, made Acetabularia an attractive model system for studies of cell biology and genetics. This umbrella-looking organism is elongated in an apical-basal direction with the root-like rhizoid in the basal end and a disc-shaped cap in the apical end, separated by a long stalk 7, 8. Acetabularia acetabulum is the most studied species of Dasycladales. Despite being unicellular, and having only a single nucleus, some species can grow to more than 10 cm in length 7. One protist group, of which we have no genomic information is the green algal order Dasycladales whose species have a very characteristic cellular morphology. 3, 4, 5, some groups are still completely devoid of any genomic information 6. Although there has been a recent increase in the number of available protist genomes e.g. This is unfortunate as protists have an enormous diversity of cellular morphologies, physiology, and genetics, possibly even more so than their multicellular relatives 2. The bulk of these genomes belong to animals, plants, and fungi, while single-celled eukaryotes (protists) remain largely absent 1. We highlight challenges associated with genome recovery and assembly of MDA data due to biases arising during genome amplification, and hope that our study can serve as a reference for future attempts on sequencing the genome from non-model eukaryotes.ĭuring the last decade, developments in DNA sequencing technology have led to a surge in the number of eukaryotic genomes being published. By amplifying and sequencing DNA from five single cells we were able to recover an estimated ~ 7–11% of the total genome, providing the first draft of the A. Here, we have attempted to overcome these challenges by amplifying genomic DNA using multiple displacement amplification (MDA) combined with microfluidics technology to distribute the amplification reactions across thousands of microscopic droplets. Since the alga has a long life cycle, is difficult to grow in dense cultures, and has an estimated diploid genome size of almost 2 Gb, obtaining sufficient genomic material for genome sequencing is challenging. However, no genomic information is available from this species. The macroscopic single-celled green alga Acetabularia acetabulum has been a model system in cell biology for more than a century.


 0 kommentar(er)
0 kommentar(er)
